Explainable Prediction of Medical Codes With Knowledge Graphs
- 1School of Information Science and Technology, Southwest Jiaotong University, Chengdu, China
- 2The Third People's Hospital of Chengdu, Chengdu, China
- 3School of Information Engineering, Chengdu University of Traditional Chinese Medicine, Chengdu, China
International Classification of Diseases (ICD) is an authoritative health care classification system of different diseases. It is widely used for disease and health records, assisted medical reimbursement decisions, and collecting morbidity and mortality statistics. The most existing ICD coding models only translate the simple diagnosis descriptions into ICD codes. And it obscures the reasons and details behind specific diagnoses. Besides, the label (code) distribution is uneven. And there is a dependency between labels. Based on the above considerations, the knowledge graph and attention mechanism were expanded into medical code prediction to improve interpretability. In this study, a new method called G_Coder was presented, which mainly consists of Multi-CNN, graph presentation, attentional matching, and adversarial learning. The medical knowledge graph was constructed by extracting entities related to ICD-9 from freebase. Ontology contains 5 entity classes, which are disease, symptom, medicine, surgery, and examination. The result of G_Coder on the MIMIC-III dataset showed that the micro-F1 score is 69.2% surpassing the state of art. The following conclusions can be obtained through the experiment: G_Coder integrates information across medical records using Multi-CNN and embeds knowledge into ICD codes. Adversarial learning is used to generate the adversarial samples to reconcile the writing styles of doctor. With the knowledge graph and attention mechanism, most relevant segments of medical codes can be explained. This suggests that the knowledge graph significantly improves the precision of code prediction and reduces the working pressure of the human coders.
Introduction
The International Classification of Diseases (ICD) is a standard classification system according to the characteristics of diseases and the rules maintained by the World Health Organization. Each code represents a specific disease, symptom, or surgery. And a set of codes in the medical record represents uniquely diagnostic and procedural information during patient visits. As a significant part of the hospital information system, it is widely used for medical insurance payments, health reports, and mortality calculations. Therefore, the ICD coding task is an essential job in the medical record information department. While ICD codes are important for making clinical and financial decisions, ICD coding is time-consuming, error-prone, and expensive. In most cases, the human coders assign ICD codes to medical records according to the clinical diagnosis record of physician. It is difficult because the code assignment should consider overall the health condition in the long text-free medical records, including symptoms, signs, surgery, medication, body, etc.
Automatic coding uses medical records as input to predict the final ICD codes based on text content. But the automatic ICD coding task usually has the following difficulties: (1) The clinical records of patients are not always structured in the same way. And the vital information in the text is distributed in various segments. For the above two reasons, it is very difficult to extract important and relevant knowledge from various kinds of medical records effectively. (2) Most importantly, the medical field has a lot of terminologies, which is difficult for non-professionals to understand the meaning of these terminologies. Even for the same disease, there are many ways to describe it differently from ICD description. (3) Datasets in the medical field are often small, and doctors have different writing styles. Each physician usually has his way to describe medical terminologies.
In this paper, we proposed a new end-to-end method called G_Coder (Graph-based Coder) for automatic ICD code assignment using clinical records. The contributions of this paper are summarized as follows: (1) We utilize Multi-CNN (multiple convolutional neural networks) to capture local correlation, which extracts key features from the irregular text. (2) We build a knowledge graph, which enriches the meaning of terminologies through integrated related knowledge points. It is combined with the attention mechanism to help understand the meaning of related terminologies, making the coding results interpretable. (3) The adversarial learning is used to generate adversarial samples to increase samples and reconcile the different writing styles.
Our model has outperformed other models in micro-AUC and micro-F1 on MIMIC-III (Multi-parameter Intelligent Monitoring in Intensive Care) datasets with 46 K distinct hospital admissions and top 50 common ICD-9 codes.
Related Works
Automatic ICD Coding
It was 20 years ago that many researchers have explored how to automatically assign ICD codes based on clinical records. There are two major categories of approaches for automatically assigning ICD-9 codes using medical records. One category is rule-based and the other category is learning-based. Rule-based systems are manually extracted statistical features by humans. Chen et al. (2017) and Ning et al. (2016) presented an improved approach based on the Longest Common Subsequence (LCS) and semantic similarity for performing ICD-10 code assignment to Chinese diagnoses. But such approaches only consider the simple matching of strings, which is not a medical problem. Beyond that, researchers applied automatic and semi-automatic (Medori and Fairon, 2010) machine learning methods to automatically assign ICD codes. Automatic ICD-9-CM encoding consisted of support vector machines (SVM) (Yan et al., 2010; Adler et al., 2011; Ferrão et al., 2013; Wang et al., 2017), k-nearest neighbors (Ruch et al., 2008; Erraguntla et al., 2012), Naive Bayes (Pakhomov et al., 2006; Medori and Fairon, 2010), and other methods such as topic model (Ping et al., 2010; Perotte et al., 2013). Semi-automatic methods generally require more manual participation and may require manual data processing, feature selection, data verification, etc. Automatic methods generally use a series of operations in an end-to-end manner. Nevertheless, the development of automatic coding technology is not yet mature, and manual verification is inevitable. All the above methods only utilize the statistical characteristics of words and ignore the contextual meaning.
In recent years, many new methods are emerging with the development of deep neural network. Li et al. (2018) combined the convolutional neural network (CNN) and the “Document to Vector” technique to extract textual features. It solves the characteristics of CNN's indistinguishable word order while taking all the words into account. Baumel et al. (2017) applied a hierarchical approach which is Hierarchical Attention bidirectional Gated Recurrent Unit (HA-GRU) to tag a discharge summary by identifying the relevant sentences. It utilizes the Gated Recurrent Unit to encode text, which experimental effect is similar to long short-term memory networks (LSTM), but it is easier to calculate. Yu Y. et al. (2019) explored character features and word features based on bidirectional LSTM with attention mechanism and Xie and Xing (2018) applied tree LSTM with ICD hierarchy information for automatic ICD coding. Compared with ordinary LSTM, bidirectional LSTMs tend to have higher accuracy, and tree LSTM is more suitable for data that is a tree-like hierarchical structure. Mullenbach et al. (2018) proposed to extract per-code textual features across the document using a convolutional neural network and used an attention mechanism to select the most relevant segments for each possible code. Based on that, Li and Yu (2019) combined multi-filter convolutional layers and residual convolutional layers to enlarge the receptive field.
Deep learning methods improved the ability to capture semantic information but ignored the importance of medical knowledge and experience. In practical work, the human coders fully utilize the basic medical knowledge to provide decision support for the work. However, all the methods just mentioned are data-driven approaches or simple mapping, which lack of the theoretical support and suffer from the complicated preprocessing of the noisy text. To build a more explainable ICD coding system, we utilize the knowledge graph as supplementary knowledge to add to the model, which is equivalent to combining a data-driven approach with medical knowledge. What is more, we successively perform text preprocessing and Multi-CNN algorithm to extract text features to reduce text noise. Adversarial learning generates adversarial samples for training to reconcile the different writing styles. The attention mechanism selects the most relevant segments for each possible code.
Graph Embedding
Graph embedding technology expresses nodes in the form of low-dimensional dense vectors, which require similar nodes in the original graph to be similar in the low-dimensional expression space. The representative work of Graph Embedding is DeepWalk (Perozzi et al., 2014), LINE (Tang et al., 2015), Node2Vec (Grover and Leskovec, 2016), SDNE (Wang et al., 2016), and Struc2Vec (Ribeiro et al., 2017). The obtained expression vectors can be used for downstream tasks, such as node classification (Ye et al., 2018; Gong and Ai, 2019), link prediction (Li et al., 2019a), or visualization (Liu et al., 2020). In the field of biomedicine, graphs are often used to predict drug interactions and predict drug target proteins. The knowledge graph embedding is used to calculate several similarity measures between all drugs in the scalable and distributed framework to obtain the interaction of drugs (Ibrahim et al., 2017). Mohamed et al. (2020) used knowledge graph embeddings to learn the vector representation of all drugs and targets to discover protein drug targets.
Attention Mechanism
The attention mechanism was first used for machine translation (Dzmitry et al., 2014). It calculates the attention weight of each word in the encoder sequence to each word in the decoder sequence to focus more on the most relevant part of the current word. The attention mechanism improves the effect and also increases the interpretability of the neural network. After adding attention, the weight of the data can be visualized to confirm the correctness of the method. Besides, attention mechanism has the ability to capture global features in long texts. The attention mechanism mimics the internal process of biological observation behavior, which is a mechanism that aligns internal experience and external sensation to increase the observation precision of some areas. It has been successfully used in medical tasks. Such as medical imaging (Ozan et al., 2018), clinical text information extraction (Li et al., 2019b; Xu et al., 2019), and DNA-related tasks (Yu W. et al., 2019; Hong et al., 2020).
Adversarial Learning
Adversarial learning is to make the two networks compete against each other. The generator network continuously captures the probability distribution of the real data in the training set and transforms the input random perturbation into new samples. The discriminator network observes both real and fake data to determine the authenticity of this data. Through repeated confrontation, the capabilities of the generator and discriminator will continue to increase until a balance is reached. Goodfellow et al. (2015) developed a method named FSGM that can effectively calculate the perturbation. They set the perturbation to the maximum value of the loss function along the direction of the gradient. FSGM takes the same step in each direction, and Goodfellow's subsequent FGM (Miyato et al., 2017) is scaled according to specific gradients to obtain better adversarial samples. Adversarial learning improves the robustness of the model through the idea of games. It randomly adds perturbation factors to the input to simulate unknown data to ensure that the model can work stably in any situation. Adversarial learning has been used for privacy protection (Max et al., 2019) of medical records and named entity recognition (Zhao et al., 2019) in clinical texts.
Materials and Methods
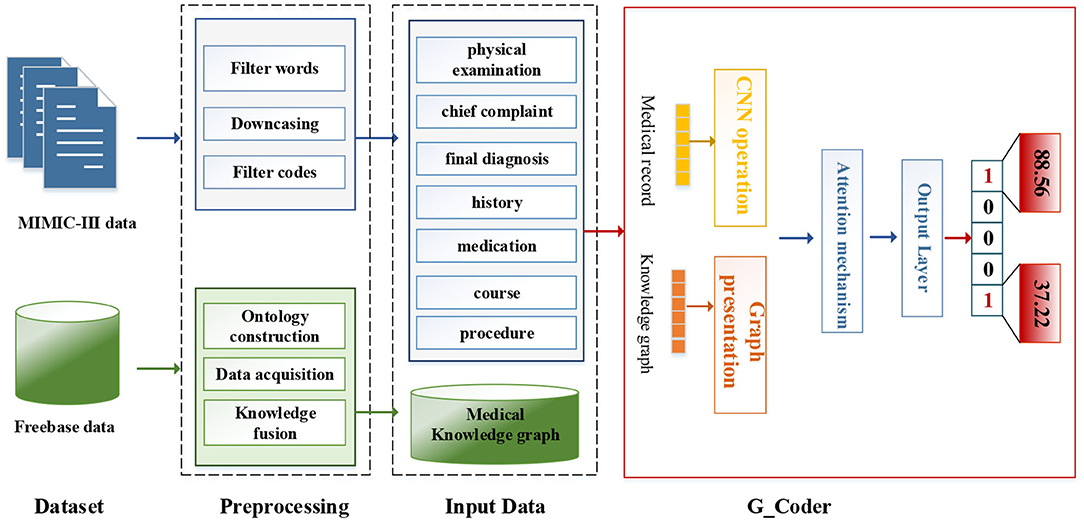
As can be seen from Figure 1, this section will detail all the processes by combining data materials with the proposed methods.
Dataset and Preprocessing
We utilize the transfer knowledge graph to improve the interpretability and performance of automatic ICD coding. In the study, we select Multi-parameter Intelligent Monitoring in Intensive Care-III (MIMIC-III) dataset (Johnson et al., 2016) as an experimental dataset and Freebase dataset as a source of the knowledge graph. A brief introduction to these two data sets and related preprocessing techniques are as follows.
MIMIC-III Dataset
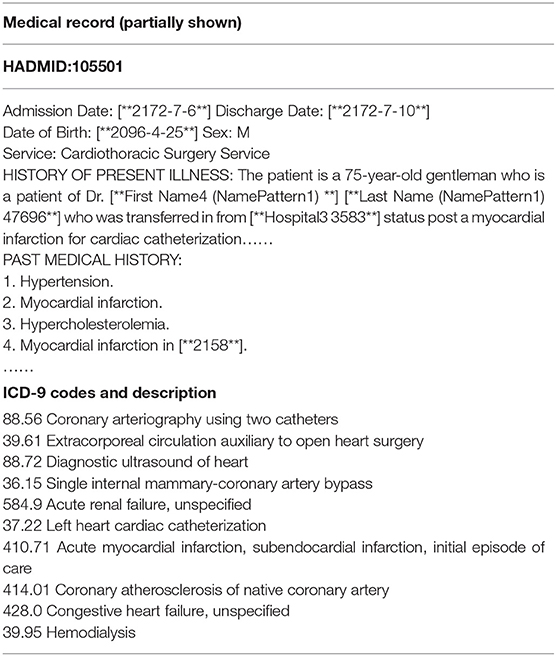
MIMIC-III dataset is the only public database for learning automated ICD-9 coding, which allows fair comparisons with different methods. It contains reliable and comprehensive 58,976 hospital admissions collected between 2001 and 2012 in the Beth Israel Deaconess Medical Center. Each medical record usually includes discharge summaries, survival data, diagnostic codes, vital signs, laboratory measurements, etc. Besides, the discharge summary always contains multiple information, such as “discharge diagnosis,” “past medical history,” physical examination,” and “chief complaint,” etc. Table 1 shows a sample of a medical record in the dataset. The “HADMID” uniquely identifies each medical record. Each hospital admission has a group of ICD-9 codes given by the medical coders. For each medical record, codes distribute unevenly in numbers which varies from one to 39. The number of codes is usually not equal to the number of diagnosis descriptions. It invalidates the one-to-one method of allocating codes. The entire dataset contains 6,984 distinct codes and 943 categories. Each code has a short phrase or a sentence, articulating a disease, symptom, or condition.
We adopt a series of standard text pre-processing techniques, which contain regular expression matching and tokenization to reduce the noise in raw note texts. Firstly, we extract relevant data from MIMIC-III as input text, which contains “physical examination,” “chief complaint,” “final diagnosis,” “history,” “medication,” “course,” and “procedure.” Secondly, we remove stop words from the input text and transform each token into its lowercase. Simultaneously removing words <3 and replacing unknown words with “UNK.” Thirdly, medical records with associated labels that do not contain the top 50 code are discarded.
Freebase
With the rapid development of the knowledge graph in recent years, research-based on knowledge graphs has attracted widespread attention in the medical field. Freebase mainly extracts structured data from wikis and publishes them as RDF. It is fully structured, but the data source is not limited to wikis. It also imports a large number of professional data sets and provides data query and entry mechanisms.
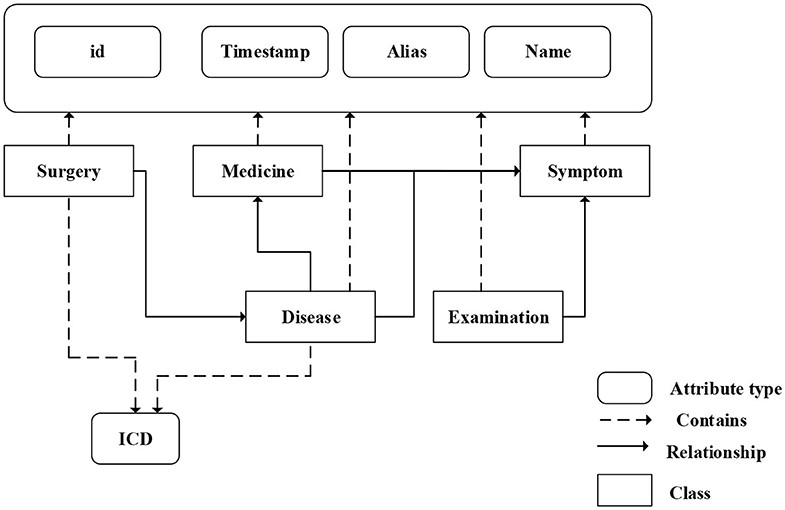
We fuse ICD-9 description information with medical knowledge extracted from freebase to build the final knowledge graph. Freebase Medicine originate from Wikipedia and other datasets such as U.S. National Medical Data. One study has reported that 70% of junior doctors used Wikipedia for health knowledge every week (Trevena, 2011). Because the freebase is reliable, the information provided in Freebase is generally considered to be reliable. The matching method is used for knowledge fusion. Since some diagnosis terms from ICD-9 description imperfectly match Freebase content, we use the ICD description text as the search terms to find the most relevant Freebase content by the Freebase API (http://freebase.gstore-pku.com/). The ontology that was constructed contains 5 entity classes, which are disease, symptom, medicine, surgery, and examination. The constructed ontology is shown in Figure 2, which contains the relationships (disease manifests as symptoms, medicine treats disease, surgery treats disease, and commonly used disease test data, etc.) and attribute types, such as id, name, ICD, etc. In the final knowledge graph, there are 1,560 nodes and more than 20,000 sets of relationships.
Methods
Overview
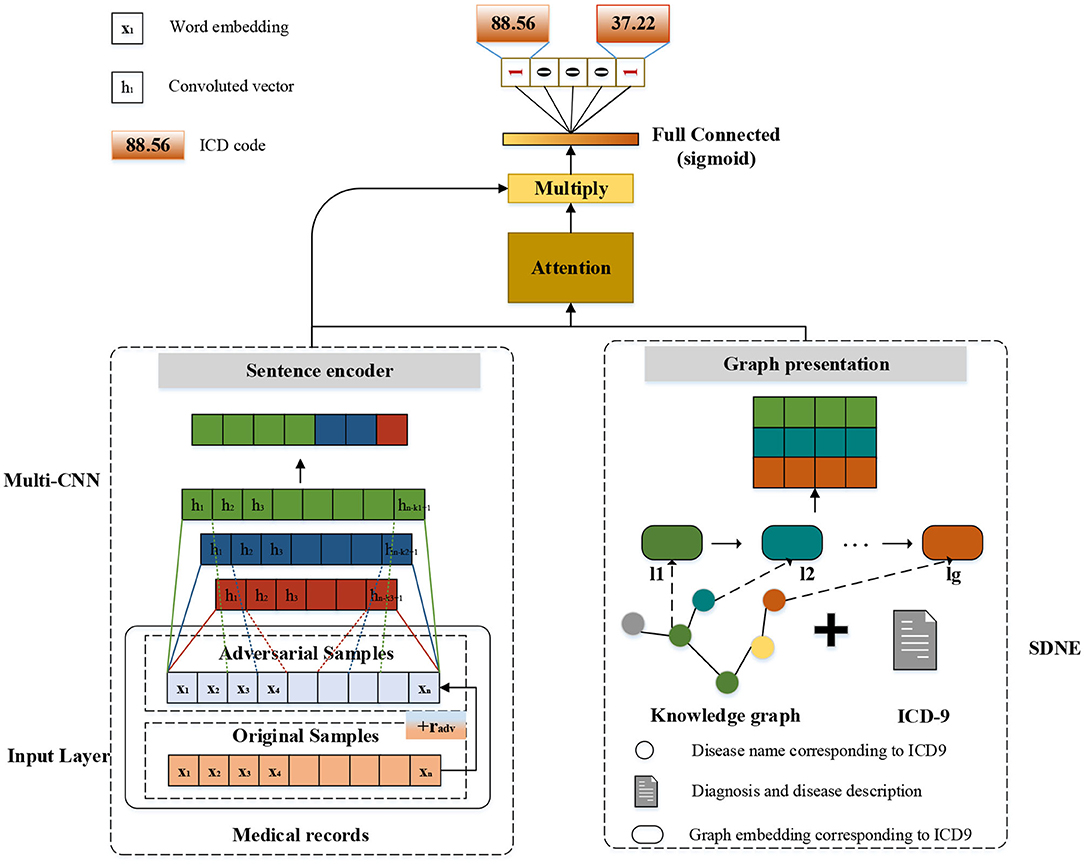
The modular method adopted in this study differs from the researchers used earlier. Figure 3 shows an overview of our approach named G_Coder. The proposed approach mainly consists of four modules, which mainly contain Multi-CNN, Graph Presentation, Attentional Matching, and Adversarial Learning.
Input Layer
Considering that the pre-trained word vectors in the medical field are not yet perfect and the experimental data in this study are very long texts, the word embeddings were initialized randomly. Leveraging a token sequence x = {x1, x2, x3…, xn} as input, where n denotes the sequence length. Assuming that the matrix W denotes the word embedding matrix, and W = {w1, w2, w3, …, wv}∈ ℝv×d, where v represents the size of total vocabulary and d represents the token dimension. The vocabulary is obtained by pre-processing the MIMIC-III clinical text. A token xi will correspond to a vector wj by looking up W. The final input of the model is a matrix X ∈ ℝn×d.
Multi-CNN
As can be seen from Figure 3, the structure of Multi-CNN is used to encode the input matrix X. Multi-CNN is a combination of multiple CNNs and MaxPooling. CNN is a kind of neural network algorithm that has successfully been applied to computer vision. MaxPooling reduces the dimension of the feature map, and effectively reduces the parameters required for subsequent layers. Besides, it magnifies the receptive field.
Multiple kernels of different sizes are used to extract key information in the sentence, which inspired by Kim (2014) who applied Text-CNN to the text classification task. Multi-CNN is used to better capture the local correlations. Assuming we have filters f1, f2, …, fm where m denotes the filter number. Each kernel size of filters denotes as k1, k2, ., km. The convolutional procedure can be formalized as formula (1),
where * denotes the convolution operator, g is an element-wise non-linear transformation, Wcm is weight parameter and bcm is the bias. Assuming that Hm = {h1, h2, h3, …, hn−k+1} is the output of m-th CNN and Hm′ is the output of m-th MaxPooling. The result of Multi-CNN is , where ⊕ denotes concatenation operator and dt denotes the dimension of Ht′.
Graph Presentation
In this study, we mainly adopt SDNE (Structural Deep Network Embedding) for medical knowledge graph node embedding. First-order proximity and second-order proximity are two crucial definitions in SDNE. The first-order proximity is used to describe the local similarity between paired nodes in the graph. If there are no directly connected edges, the first-order proximity is 0. The second-order proximity measures the similarity of their neighbor sets between two nodes. The optimization goal of SDNE is shown in formulas (2–4):
Each si contains the neighbor structure information of the i-th node. The letter r denotes the vector representation of each node. Where nd denotes the number of neighbors at nodes i.
L1st makes the embedding vectors corresponding to the two adjacent nodes in the graph close in the hidden space. Lreg is a regularization constraint, α is a parameter that controls the first- proximity loss, and β is a parameter that controls the regularization constraint. After SDNE, each node gets its own vector representation in the hidden space. Assuming that the matrix yg is the result linked to ICD-9 of SDNE, which . Where lg denotes the number of ICD-9 and dg denotes the dimensions of each node.
Attentional Matching
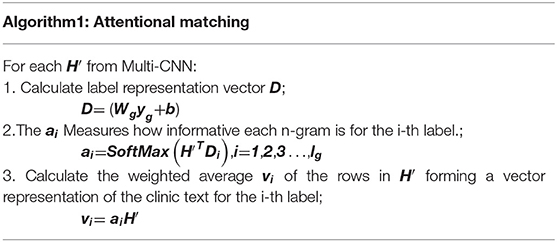
Human coders usually look for the most critical part of the medical record (Such as symptoms, complications, etc.) to determine the final coding result. In this task, we need to refine the text that most relevant to the ICD information and give higher weight. For the above reasons, we apply the attention mechanism. A benefit is that it selects the segments from the text that are most relevant to each predicted label. The specific algorithm details are shown in Table 2. It obtained the clinical text representation vector H′ through preprocessing and Multi-CNN, and at the same time obtained the ICD coded representation yg using the knowledge graph embedding results. A linear transformation was performed on the code representation to obtain the final code representation D, which has the same dimensions as the number of codes. The text representation H′ and label representation D are used to calculate the weight ai of the relationship between each label and each segment of the text. Finally, the text H′ and weight ai are used to weight the average of each part of the text to obtain the final clinical text representation vi.
The results in Table 2 can be summarized as follows:
Where , and exp is an exponential function with natural constant e as a base. The matrix is the weight parameter. And A denotes attention weights for each pair of an ICD code and the text. The letter denotes the output of the attention. The concrete example can be found in Table 7.
Adversarial Learning
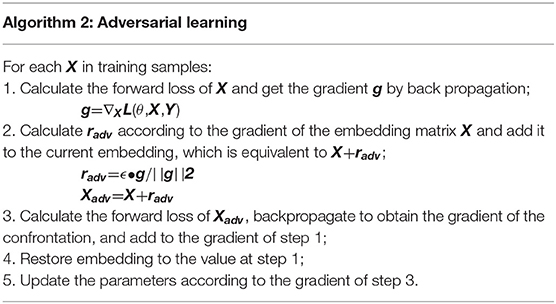
We apply FGM (fast gradient method) to reconcile the different writing styles of doctors and increase training samples (Miyato et al., 2017). The basic idea is: The writing of medical records follows the writing standards, but also contains different writing styles. Adversarial learning weakens the influence of writing style. The purpose of adversarial training is that the model will work steadily even if there are large differences in doctor writing styles. FGM uses a first-order Taylor expansion on the adversarial objective function to approximate to maximize the error output by the model, which is equivalent to using a single-step gradient descent method with a step size of ϵ to find the adversarial samples. The specific algorithm details are shown in Table 3. It calculates the gradient g of the clinic text embedding X after forward propagation and then back propagation. The gradient is used to calculate the perturbation radv added to X. After such a process, Xadv is an automatically generated adversarial sample. It uses the adversarial samples to calculate together with the original samples, increasing the number of samples, while mimicking the writing style of different doctors.
The goals of adversarial learning are as follows:
The formula (7) is divided into two parts, one is the maximization of the internal loss function, and the other is the minimization of the external risk. In the internal max, L is the defined loss function, D is the perturbation of input samples, and R is the space for a perturbation. The goal of adversarial learning is to find the amount of perturbation that makes the most judgment errors. For the above attacks, the most robust model parameters are found. After further optimizing the model parameters, the expected value of the entire data distribution is still minimal.
Output Layer
We compute a probability for label vector using full connection layer and a sigmoid transformation by the output of attention representation V:
Where is learnable weights of output layer and . The whole learning process minimizes the binary cross-entropy loss (9) of prediction probability and the target Yi ∈ (0, 1). The label i is selected when is >0.5.
Where X denotes the input word sequence, λ is the L2 regularization hyperparameter. And θ denotes all the parameters. We utilize the back-propagation algorithm and Adam optimizer (Kingma and Ba, 2014) to train the model.
Experiments
Experimental Settings
A majority of codes are only assigned to too few medical records. Since the top 50 common ICD-9 codes covered 93.6% of the all dataset, we pick 50 most frequent codes to carry out the experiment while considering that our method can readily be extended to more codes as long as sufficient training data is available. The experimental dataset using top-50 codes has a total of 46,552 discharge summaries, which has 43,000 discharge summaries for training, 1,800 for validation, and 1,752 for the test. In this experiment, the settings are shown in Table 4. The token dimension d is 100; the knowledge graph embedding size dg is 128; the out-channel size df of a filter in the Multi-CNN layer is 50; the learning rate lr is 0.001; the L2 regularization hyperparameter λ is 0.00001; the max length of each medical record is 1,800; the mini-batch size is 16 and the dropout rate dp is 0.4. We used three filters and the kernel size of filters is 4,5,6.
Evaluation Metrics
This task can be regarded as a multi-label classification problem. Therefore, we evaluate the method by micro−F1 and AUC (Area under the curve). The micro−F1 is harmonic mean that calculated from Precision and Recall. All evaluation matrixes are calculated as follows:
In these formulas, TPi is the set of ground truth labels of each class, n is the number of samples, FNi is the number of positive classes predicted as negative classes and FPi is the number of negative classes predicted as positive classes. AUC is mainly used to evaluate the ranking ability of the current model. The higher the AUC, the better the ranking ability of the model. When the prediction probability values of all positive samples are higher than the negative samples, the AUC of the model is 1.
Results
Model Comparison
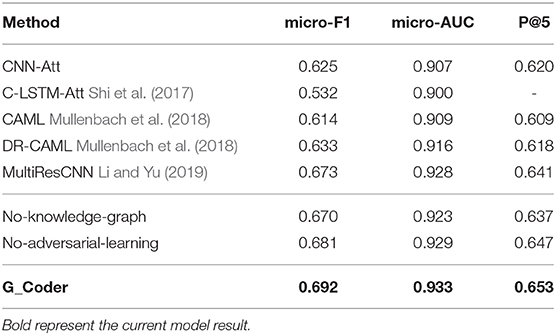
This section illustrates the performance of our approach. The experimental results of the top-50 codes show in Table 5, which show that our work has improved on previous work. CNN-Att is the baseline model for this experiment, which uses CNN to encode text. MultiResCNN has achieved the state-of-the-art results on the MIMIC-III datasets using unstructured text. Besides, their work is based on CAML and the model is improved. It mainly consists of a multi-filter convolutional layer and residual convolutional layer for multi-label classification. C-LSTM-Att applied LSTM-based language models to encode clinical notes and ICD codes and applied an attention method to solve the mismatch between clinical notes and codes. They focused on predicting the 50 codes that have the top frequencies for the medical records in the MIMIC-III dataset just like us.
Comparing our model with existing work for automatic ICD coding. As shown in Table 5, the conclusions are as follow:
1) G_Coder obtains better results in the micro-AUC, micro-F1, and P@5. Compared with the state-of-the-art model MultiResCNN, G_Coder improves the micro-AUC by 0.005, the micro-F1 by 0.019, the P@5 by 0.012. P@5 measures the ability of the method to return the top 5 high-confidence subsets of codes. Our approach achieves relatively high precision of the five most confident predictions, on average 3.3 are correct.
2) CNN-based models are more suitable for this task. LSTM pay more attention to capture long sequence features, and cannot extract important local features from noise text. Simultaneously, the length of the medical record text makes the recurrent neural network have extremely high requirements for machine performance in this task. In contrast, it can be seen from the model construction that CNN can better extract long text features, and multilayer CNN with different convolution kernels can better capture local correlation.
3) The attention mechanism is essential. Each model utilizes the attention mechanism, which shows that the mechanism accurately highlights the information related to ICD in the text. The following content will prove the value of the knowledge graph and adversarial learning in this task.
Ablation Study
To gain more insight, the ablation study applied to verify the effectiveness of the adversarial learning and knowledge graph. To evaluate each module, we perform single variable experiments. The comparisons of the No-one module with the full model are given in Table 6. We remove one module from the full model without changing other modules and denote such a baseline by No-X. To evaluate them, we compared with the two configurations: (1) No- knowledge-graph, which removes the graph presentations and directly uses a randomly initialized vector as final representations of codes information; (2) No- adversarial-learning, which removes the adversarial learning form full model.
It can see from Table 6 that our full model obtains better results in all evaluation matrix. Compared with the full model, No-knowledge-graph dropped the micro-AUC from 0.933 to 0.923, the micro-F1 from 0.692 to 0.670, the P@5 from 0.653 to 0.637. At the same time, No-adversarial-learning dropped the micro-AUC from 0.933 to 0.929, the micro-F1 from 0.692 to 0.681, the P@5 from 0.653 to 0.647. The above results show that the knowledge graph-based method can add clinical experience to make the results better. And adversarial learning generates adversarial samples through perturbation factors to enhance the generalization ability of the model on the test set. From the results we have obtained, one can conclude that the combination of data-driven and medical knowledge can enhance the precision of ICD automatic coding.
Universality Study
To prove that the knowledge graph is universal in this task. We design the experiment, which is to add a knowledge graph to the basic baseline model and compare it with the baseline model.
According to the experimental results in Table 6, it can be seen that the knowledge graph not only performs well in G_Coder but also can be extended to other model structures. The knowledge graph improves the micro-F1 of the baseline model by 2.6%. This shows that the knowledge graph is universal and can be flexibly grafted into other model structures.
Evaluation of Interpretability
We use two methods to verify the interpretability. The first is an intuitive method that attention extracts keywords and displays the correlation between the code and the evidence. Examples can be found in Table 7. It can be seen from which words the basis of coding comes from. Taking 584.9 as an example, there is an information overlap between “acute renewal failure, unspecified” and “with acute renewal failure secondary” in clinical texts.
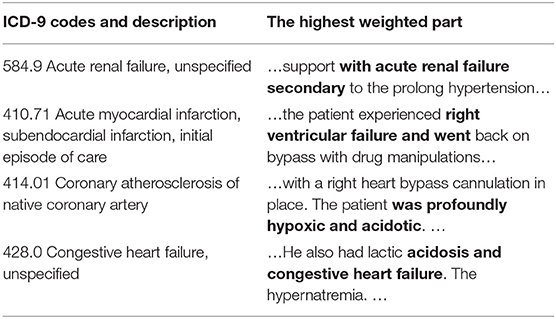
Table 7. Presentation of clinical text fragments and their corresponding ICD codes (The bold part indicates the highest weight).
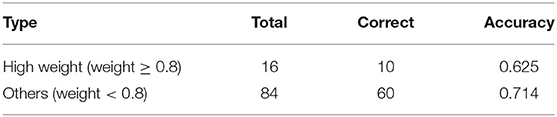
The second is a quantitative method where doctors judge the results of attention distribution. A clinical medical record was randomly selected, and segments were extracted based on the results of its attention. We select 5-words in this setting to emulate a span of attention over words likely to be given by a human reader. Since the segment may overlap, the most important 5-words were extracted according to attention weight. As can be seen from Table 8, the score is divided into two stages, one is high weight, that is >0.8, and the other is <0.8. In a total of 100 segments, there are 16 with a weight >0.8 and 84 with a weight <0.8. According to the evaluation results of human coders, 10 of the high weights are correct, and the remaining correct number is 60.
Conclusions and Discussions
Conclusions
Inspired by the structure of graphs that can model the relationships and knowledge between all things in the world, we think the graph structure can connect the parts of the data in this task and create a knowledge graph using medical-related data from the Freebase database. At the same time, the development of deep learning has also allowed further development of natural language processing such as automatic coding and text classification. In this paper, we propose a new explainable method for automatic ICD coding. The result of the micro-F1 score of 50 most frequent codes is 69.2%, which outperforms all the other models especially when raw clinical text data is used as input features to the prediction models.
The experimental evaluation of the MIMIC-III dataset shows the following points. First, we combined deep learning with knowledge graphs in ICD coding tasks. The medical knowledge graph supervises the coding process as a teacher. At the same time, we apply the SDNE algorithm to encode each entity of the knowledge graph and link it to the ICD-9 code. The Multi-CNN algorithm is utilized to encode long text information of MIMIC-III data. In the attention mechanism, we combine the two mentioned above to identify the segments of text that are most relevant to each ICD-9 code. Finally, we generate adversarial samples through adversarial training and send the samples to the training along with the original samples. It can weaken the influence of writing style and make model more stable. Moreover, in the ablation study and universality study, we use the single variable rule to verify the importance of adversarial learning and knowledge graph. The results prove that the knowledge graph can be flexibly grafted into the model structure to help understand the terminology. Two methods are used to verify the interpretability of the method. It is confirmed that this method is based on the important basis in the clinical text for ICD coding. G_Coder has a higher accuracy rate than the other method. And before the coder works, G_Coder can perform ICD pre-selection to save time for whole encoding work.
Discussions
The major limitation of this work is that it does not perform well on infrequent codes. To achieve fully automatic coding, infrequent coding has to be considered. And we hold that the method can readily be extended to more codes as long as sufficient training data is available. In addition, the new ICD version should also be considered, such as ICD10, ICD11, etc. ICD classification is a disease classification directory with hierarchical relationship. The structure of ICD is also a direction worth considering.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://freebase.gstore-pku.com/, https://mimic.physionet.org/, https://developers.google.com/freebase.
Author Contributions
WY and FT provided total research ideas designed the experiments. WY performed the experiments and wrote the first draft of the manuscript. LH and LC guided the experiment as experts and analyzed the results. FT contributed to the High-performance experimental equipment. WY and QX contributed to manuscript revision, reading, and approving the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Sichuan Science and Technology Program (No. 2017SZYZF0002), National Key R&D Program of China (No. 2019YFB2101802), and Sichuan Key R&D project (No. 2020YFG0035).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Beth Israel Deaconess Medical Center for providing data support. We would also like to thank the reviewers for their insightful comments.
References
Adler, J. P., Frank, W., Noemie, E., and Nicholas, B. (2011). Hierarchically Supervised Latent Dirichlet Allocation. Advances in Neural Information Processing Systems, 2609–2617. Available online at: http://papers.nips.cc/paper/4313-hierarchically-supervised-latent-dirichlet-allocation
Baumel, T., Nassour-Kassis, J., Elhadad, M., and Elhadad, N. (2017). Multi-Label Classification of Patient Notes a Case Study on ICD Code Assignment. arXiv. Available online at: https://arxiv.org/abs/1709.09587 (accessed September 27, 2017).
Chen, Y., Lu, H., and Li, L. (2017). Automatic ICD-10 coding algorithm using an improved longest common subsequence based on semantic similarity. PLoS ONE. 12:e0173410. doi: 10.1371/journal.pone.0173410
Dzmitry, B., Kyunghyun, C., and Yoshua, B. (2014). Neural Machine Translation by Jointly Learning to Align and Translate. arXiv. Available online at: https://arxiv.org/abs/1409.0473 (accessed September 1, 2014).
Erraguntla, M., Gopal, B., Ramachandran, S., and Mayer, R. (2012). “Inference of missing ICD 9 codes using text mining and nearest neighbor techniques,” in 2012 45th Hawaii International Conference on. IEEE (HICSS), 1060–1069. doi: 10.1109/HICSS.2012.323
Ferrão, J., Janela, F., Oliveira, M., and Martins, H. (2013). “Using structured EHR data and SVM to support ICD-9-CM coding,” in Proceedings of the 2013 IEEE International Conference on Healthcare Informatics (Philadelphia, PA), 511–516. doi: 10.1109/ICHI.2013.79
Gong, P., and Ai, L. (2019). Neighborhood Adaptive Graph Convolutional Network for Node Classification. New Jersey, NJ: IEEE Access, 170578–170588. doi: 10.1109/ACCESS.2019.2955487
Goodfellow, I., Shlens, J., and Szegedy, C. (2015). Explaining and Harnessing Adversarial Examples. Available onlibe at: https://arxiv.org/abs/1412.6572 (accessed March 20, 2015).
Grover, A., and Leskovec, J. (2016). “node2vec: scalable feature learning for networks,” in KDD: Proceedings. International Conference on Knowledge Discovery & Data Mining (San Francisco, CA), 855–864. doi: 10.1145/2939672.2939754
Hong, Z., Zeng, X., Wei, L., and Liu, X. (2020). Identifying enhancer–promoter interactions with neural network based on pre-trained DNA vectors and attention mechanism. Bioinformatics 36, 1037–1043. doi: 10.1093/bioinformatics/btz694
Ibrahim, A., Achille, F., Oktie, H., Ping, Z., and Mohammad, S. (2017). Large-scale structural and textual similarity-based mining of knowledge graph to predict drug–drug interactions. J. Web Sem. 44, 104–117. doi: 10.1016/j.websem.2017.06.002
Johnson, A. E. W., Pollard, T. J., Shen, L., Lehman, L., Feng, M., Ghassemi, M., et al. (2016). MIMIC-III, a freely accessible critical care database. Scient. Data 3:160035. doi: 10.1038/sdata.2016.35
Kim, Y. (2014). “Convolutional neural networks for sentence classification,” Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (Doha), 1746–1751. doi: 10.3115/v1/D14-1181
Kingma, D., and Ba, J. (2014). “Adam: a method for stochastic optimization,” in International Conference on Learning Representations. Available online at: https://arxiv.org/abs/1412.6980 (accessed December 22, 2014).
Li, F., and Yu, H. (2019). “ICD coding from clinical text using multi-filter residual convolutional neural network,” in AAAI Technical Track: Natural Language Processing (New York, NY), 34. doi: 10.1609/aaai.v34i05.6331
Li, M., Fei, Z., Zeng, M., Wu, F., Li, Y., Pan, Y., et al. (2018). “Automated ICD-9 coding via a deep learning approach,” in IEEE/ACM Transactions on Computational Biology and Bioinformatics (New Jersey, NJ), 16, 1193–1202. doi: 10.1109/TCBB.2018.2817488
Li, Z., Liu, Z., Huang, J., Tang, G., Duan, Y., Zhang, Z., et al. (2019a). MV-GCN: multi-view graph convolutional networks for link prediction. IEEE Access 7, 176317–176328. doi: 10.1109/ACCESS.2019.2957306
Li, Z., Yanga, J., Goua, X., and Qi, X. (2019b). Recurrent neural networks with segment attention and entity description for relation extraction from clinical texts. Artif. Intell. Med. 97, 9–18. doi: 10.1016/j.artmed.2019.04.003
Liu, H., Li, Y., Hong, R., Li, Z., Li, M., Pan, W., et al. (2020). Knowledge graph analysis and visualization of research trends on driver behavior. J. Intell. Fuzzy Syst. 38, 495–511. doi: 10.3233/JIFS-179424
Max, F., Arne, K., Gregor, W., and Chris, B. (2019). “Adversarial learning of privacy-preserving text representations for de-identification of medical records,” in Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics (Florence), 5829–5839.
Medori, J., and Fairon, C. (2010). “Machine learning and features selection for semi-automatic ICD-9-CM encoding,” Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents (Los Angeles, CA), 84–89.
Miyato, T., Dai, A., and Goodfellow, I. (2017). Adversarial Training Methods for Semi-Supervised Text Classification. Available online at: https://arxiv.org/abs/1605.07725 (accessed May 6, 2017).
Mohamed, S. K., Nováček, V., and Nounu, A. (2020). Discovering protein drug targets using knowledge graph embeddings. Bioinformatics. 36, 603–610. doi: 10.1093/bioinformatics/btz600
Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., and Eisenstein, J. (2018). Explainable prediction of medical codes from clinical text. NAACL 1, 1101–1111. doi: 10.18653/v1/N18-1100
Ning, W., Yu, M., and Zhang, R. (2016). A hierarchical method to automatically encode Chinese diagnoses through semantic similarity estimation. BMC Med. Inform. Dec. Making. 16, 1–12. doi: 10.1186/s12911-016-0269-4
Ozan, O., Jo, S., Loic, L. F., Matthew, L., Mattias, H., Kazunari, M., et al (2018). Attention U-Net: Learning Where to Look for the Pancreas. Available online at: https://arxiv.org/abs/1804.03999 (accessed April 11, 2018).
Pakhomov, S., Buntrock, J., and Chute, C. (2006). automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. J. Am. Med. Inform. Assoc. 13, 516–525. doi: 10.1197/jamia.M2077
Perotte, A., Pivovarov, R., Natarajan, K., Weiskopf, N., Wood, F., and Elhadad, N. (2013). Diagnosis code assignment: models and evaluation metrics. JAMIA 21, 231–237. doi: 10.1136/amiajnl-2013-002159
Perozzi, B., Al-Rfou, R., and Skiena, S. (2014). “DeepWalk: online learning of social representations,” in Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (New York, NY). doi: 10.1145/2623330.2623732
Ping, C., Araly, B., and Chris, R. (2010). “Semantic analysis of free text and its application on automatically assigning ICD-9-CM codes to patient records,” in Proceedings of the 9th IEEE International Conference on Cognitive Informatics (ICCI) (Beijing), 68–74.
Ribeiro, L., Saverese, P., and Figueiredo, D. (2017). “struc2vec: Learning node representations from structural identity,” Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (Halifax, NS), 385–394. doi: 10.1145/3097983.3098061
Ruch, P., Gobeill, J., Tbahritia, I., and Geissbühlera, A. (2008). “From episodes of care to diagnosis codes: automatic text categorization for medico-economic encoding,” in AMIA. Annual Symposium Proceedings/AMIA Symposium (Washington, DC: AMIA Symposium), 636–640.
Shi, H., Xie, P., Hu, Z., Zhang, M., and Xing, E. (2017). Towards Automated ICD Coding Using Deep Learning. Available at: https://arxiv.org/abs/1711.04075 (accessed November 11, 2017).
Tang, J., Qu, M., Wang, M., Zhang, M., Yan, J., and Mei, Q. (2015). LINE: “Large-scale information network embedding,” in WWW '15: Proceedings of the 24th International Conference on World Wide Web (Florence), 1067–1077. doi: 10.1145/2736277.2741093
Wang, D., Cui, P., and Zhu, W. (2016). “Structural deep network embedding,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: KDD (San Francisco, CA), 1225–1234. doi: 10.1145/2939672.2939753
Wang, S., Li, X., Chang, X., Yao, L., Sheng, Q., and Long, G. (2017). Learning multiple diagnosis codes for ICU patients with local disease correlation mining. ACM Trans. Knowl. Disc. Data. 11, 1–21. doi: 10.1145/3003729
Xie, P., and Xing, E. (2018). “A neural architecture for automated ICD coding,” in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Melbourne, VIC), 1066–1076. doi: 10.18653/v1/P18-1098
Xu, K., Yang, Z., Kang, P., Wang, Q., and Liu, W. (2019). Document-level attention-based BiLSTM-CRF incorporating disease dictionary for disease named entity recognition. Comp. Biol. Med. 108, 122–132. doi: 10.1016/j.compbiomed.2019.04.002
Yan, Y., Fung, G., Dy, J., and Rosales, R. (2010). “Medical coding classification by leveraging inter-code relationships,” in Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (Washington, DC), 193–202. doi: 10.1145/1835804.1835831
Ye, Q., Zhu, C., Li, G., Liu, Z., and Wang, F. (2018). Using node identifiers and community prior for graph-based classification. Data Sci. Eng. 3, 68–83. doi: 10.1007/s41019-018-0062-8
Yu, W., Yuan, C., Qin, X., Huang, Z. H., and Li, S. (2019). “Hierarchical attention network for predicting DNA-protein binding sites,” in Proceedings of the International Conference on Intelligent Computing (ICIC) (Nanchang), 11644, 366–373. doi: 10.1007/978-3-030-26969-2_35
Yu, Y., Li, M., Liu, L., Fei, Z., Wu, F. X., and Wang, J. X. (2019). Automatic ICD code assignment of chinese clinical notes based on multilayer attention BiRNN. J. Biomed. Inform. 91:103114. doi: 10.1016/j.jbi.2019.103114
Keywords: automated ICD coding, knowledge graphs, explainable, medical records, natural language processing
Citation: Teng F, Yang W, Chen L, Huang L and Xu Q (2020) Explainable Prediction of Medical Codes With Knowledge Graphs. Front. Bioeng. Biotechnol. 8:867. doi: 10.3389/fbioe.2020.00867
Received: 19 April 2020; Accepted: 06 July 2020;
Published: 14 August 2020.
Edited by:
Yungang Xu, University of Texas Health Science Center at Houston, United StatesReviewed by:
Sijia Liu, Mayo Clinic, United StatesErick Antezana, Norwegian University of Science and Technology, Norway
Copyright © 2020 Teng, Yang, Chen, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Teng, fteng@swjtu.edu.cn
 Fei Teng1*
Fei Teng1*  Wei Yang
Wei Yang